Int. J. Dev. Biol. 66: 85 - 95 (2022)
Special Issue: Developmental Biology in Greece
Promyelocytic leukemia protein (PML) and stem cells: from cancer to pluripotency
Review | Published: 26 October 2021
Abstract
The promyelocytic leukemia protein (PML) is the core organizer of cognate nuclear bodies (PML-NBs). Through physical interaction or modification of diverse protein clients, PML-NBs regulate a multitude of - often antithetical- biological processes such as antiviral and stress response, inhibition of cell proliferation and autophagy, and promotion of apoptosis or senescence. Although PML was originally recognized as a tumor-suppressive factor, more recent studies have revealed a “double-faced” agent role for PML. Indeed, PML displayed tumor cell pro-survival and pro-migratory functions via inhibition of migration suppressing molecules or promotion of transforming growth factor beta (TGF-β) mediated Epithelial-Mesenchymal Transition (EMT) that may promote cancer cell dissemination. In this line, PML was found to correlate with poor patient prognosis in distinct tumor contexts. Furthermore, in the last decade, a number of publications have implicated PML in the physiology of normal or cancer stem cells (CSCs). Promyelocytic leukemia protein activates fatty acid oxidation (FAO), a metabolic mechanism required for the asymmetric divisions and maintenance of hematopoietic stem cells (HSCs). In embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), PML is required for maintenance of the naïve and acquisition of the induced pluripotency state, respectively. Correspondingly, PML ablation causes significant morphological gene expression and lineage choice changes. In this review, we focus on the mechanisms orchestrated by PML and PML-NBs in cancer and healthy stem cells, from cell physiology to the regulation of chromatin dynamics.
Keywords
PML, stem cells, cancer, pluripotency, epigenetics
Introduction - PML and PML Bodies
The Promyelocytic Leukemia (PML) gene was first described in the early 1990s at the point of chromosomal translocation t (15, 17), where it was found to encode an oncogenic chimeric protein emerging from the fusion of PML and the retinoic acid receptor a (RARa) in patients with acute promyelocytic leukemia (APL). The PML-RAR protein prevents the induction of transcription by RARα and disrupts the PML nuclear bodies (PML-NBs) formed by PML (see below in introduction), eventually blocking the differentiation of bone marrow progenitor cells, sustaining their self-renewal capacity and preventing their apoptotic process (Kakizuka et al. 1991; de Thé et al. 1991).
The PML protein belongs to a family of 70 proteins in humans, which are characterized by the presence of a tripartite motif family (TRIM) and a RBCC (RING finger/B box/coiled-coil) region, which is conserved in all PML isoforms (Borden, 2008). Promyelocytic leukemia protein is the key component of PML-NBs, which are nuclear protein structures self-assembled through the RBCC motif interactions. PML nuclear bodies can be found in the nucleoplasm, nucleolus, the nuclear envelope and the cytoplasm. The PML gene contains nine exons that, by alternative splicing or internal initiation, allow for the generation of seven basic protein isoforms (PMLI to PMLVII). Additional putative isoforms are thought to be produced (https://genome.ucsc.edu/). Most PML protein isoforms bear a SUMO-interacting motif (SIM) for binding to sumoylated proteins. Isoforms PMLI to PMLVI bear a Nuclear Localization Signal (NLS) that allows for nuclear import. PMLI has a Nuclear Export Signal (NES) that allows the protein to shuttle between the nucleus and cytoplasm, in contrast with PMLVII isoform, which lacks the NLS motif and is only found in the cytoplasm (Shen et al. 2006; Nisole et al. 2013; Buczek et al. 2016). The PML protein is a sensor of cellular stress and other environmental signals, including growth factors and cytokines, and its abundance is regulated transcriptionally and post-translationally in response to cellular stress.
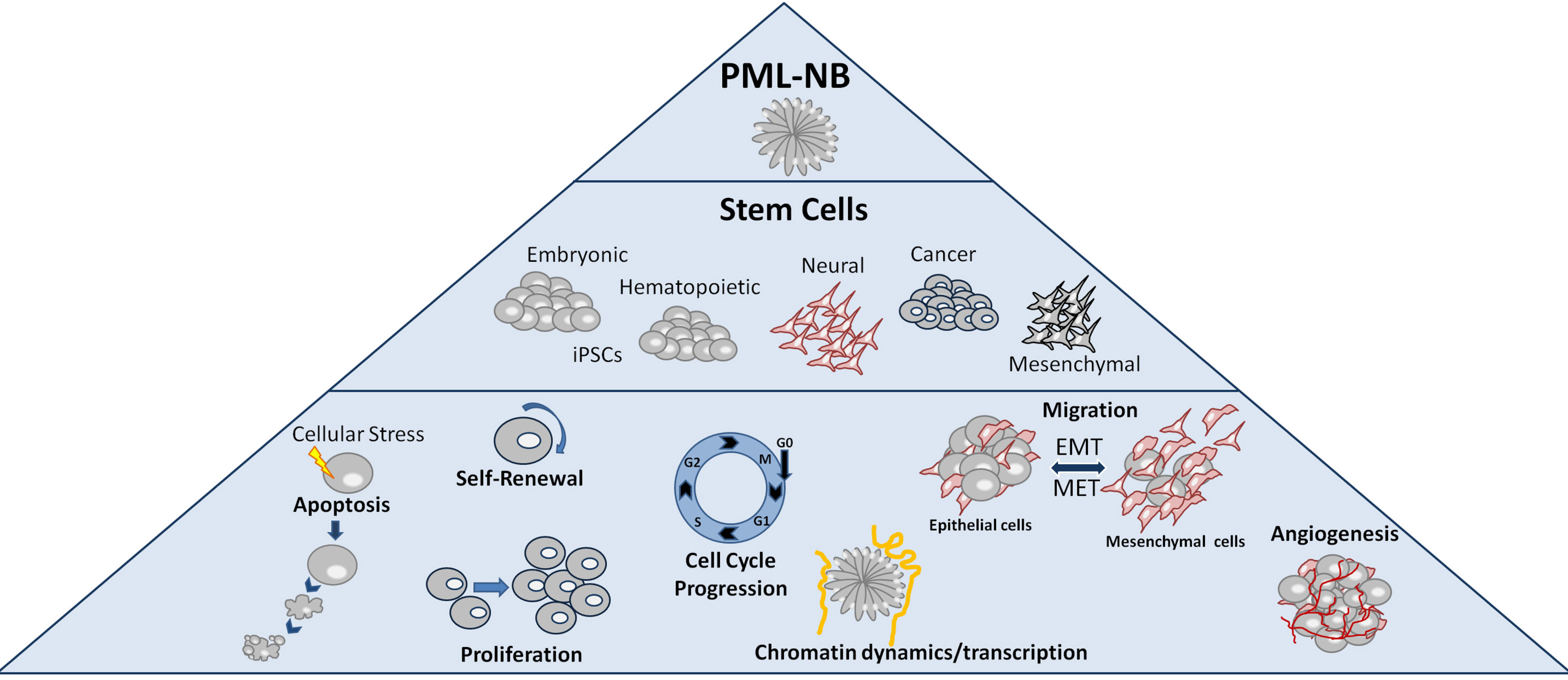
PML nuclear bodies, originally called Kremer bodies, were named after the PML protein that is their key component. They are distinct nuclear membrane-less spherical structures, 0.2 to 1.0μm in size, found in most mammalian cells, typically at about 1 to 30 particles per nucleus, depending on the cell type, cell cycle phase and cell differentiation state (Bernardi and Pandolfi, 2007). PML-NBs formation is based on the biophysical mechanism of liquid-liquid phase separation (Corpet et al. 2020), according to which concentrated droplets are form in the nucleoplasm when the local concentration of biomolecules exceeds a threshold. The interactions between SUMO and SIM in PML-NBs create such droplets within the cell nucleus (Banani et al. 2016; Uversky, 2017). By interacting and/or participating in the post-translational modifications of various nuclear proteins, often referred to as client-proteins (Van Damme et al. 2010), PML-NBs regulate a variety of cellular processes such as apoptosis, proteolysis, cellular aging, cell self-renewal capacity, DNA damage response, telomere stability, gene expression and chromatin/epigenetic states (Hoischen et al. 2018; Hsu and Kao, 2018; Lallemand-Breitenbach and de Thé, 2018). In this review, we focus on the diverse functions that are regulated by PML and PML-NBs in normal/healthy and cancer stem cells. We also discuss recently emerged roles for PML-NBs in Epithelial-Mesenchymal Transitions (EMT), chromatin dynamics and transcriptional regulation (Fig. 1).
The functions of PML-NBs in Cancer and Stem cells
Cancer and Cancer Stem Cells
Cancers are characterized by a molecular, cellular and phenotypic heterogeneity. A minor tumor cell population has the ability to self-renew and partly differentiate, thus helping to maintain or increase the tumor. Because they share common properties with normal tissue stem cells, they were termed cancer stem cells (CSCs) and were initially identified in leukemias and later in solid tumors (Beck and Blanpain, 2013). Cancer stem cells survive traditional cancer treatment, such as drugs or radiotherapy, are able to repopulate the tumor, and thus are responsible for cancer relapse and metastasis. Cancer stem cells are considered to be the most promising targets for cancer treatment (Yang et al. 2020). Embryonic stem cells (ESCs) and cancer stem cells share common characteristics such as rapid proliferation, similar metabolic requirements and self-renewal capacity. In addition, mechanisms of mesenchymal and epithelial transitions (EMT & MET) are involved in both somatic cell reprogramming to pluripotency and cancer stem cell migration in metastases. Interestingly, the CSC and ESC phenotypes seem to be governed by common transcription factors (Oct4, Nanog, Sox2, Klf4 and c-Myc), signaling pathways (Wnt, Notch, Hedgehog and TGF/SMAD) and epigenetic mechanisms (Hadjimichael et al. 2015). Moreover, the tumor microenvironment (TME) protects CSCs from genotoxic insults and increases their chemical and radiological resistance. The tumor microenvironment is composed of cancer-associated fibroblasts and mesenchymal stem cells, other adjacent tissue cells, micro-vessels, immune cells and their various secreted regulatory factors. Signals from the TME have a major role in tumor progression. Conversely, tumor secreted proteins or exosomes may promote angiogenesis, induce differentiation of tumor-related fibroblasts, participate in immune regulation and condition the pre-metastatic niche. Furthermore, within the TME, the hypoxic environment of the TME promotes CSCs self-renewal, cancer cell invasion and metastasis, through the major hypoxia response factor 1a (HIF1A) (Yang et al. 2020). The acidic microenvironment around hypoxic cells favors the activation of proteases that contribute to metastasis. Finally, hypoxic cells are less likely to respond to chemotherapeutics, due to local abnormal angiogenesis and the inaccessibility in their niche. As a result, eliminating CSCs by targeting the hypoxic CSC niche in combination with chemotherapy may be a viable option (Carnero and Lleonart, 2016).
Although no PML translocations, such as those described in APL, occur in solid tumors, early studies correlated their lack of expression in samples of primary tumors, including breast cancer (Gurrieri et al. 2004). Promyelocytic leukemia protein contributes to activation of the apoptotic process and cellular aging by stabilizing the p53 protein (Ferbeyre et al. 2000; de Stanchina et al. 2004), It also acts as a tumor suppressor through the PI3K/ Akt/mTOR interaction at multiple levels and suppresses the migratory capacity of breast and endothelial cancer cells, while it seems to negatively regulate HIF mediated angiogenesis (Bernardi et al. 2006; Trotman et al. 2006; Reineke et al. 2008, 2010; Scaglioni et al. 2012; Yamada et al. 2014; Hsu et al. 2016; Hsu et al. 2017).
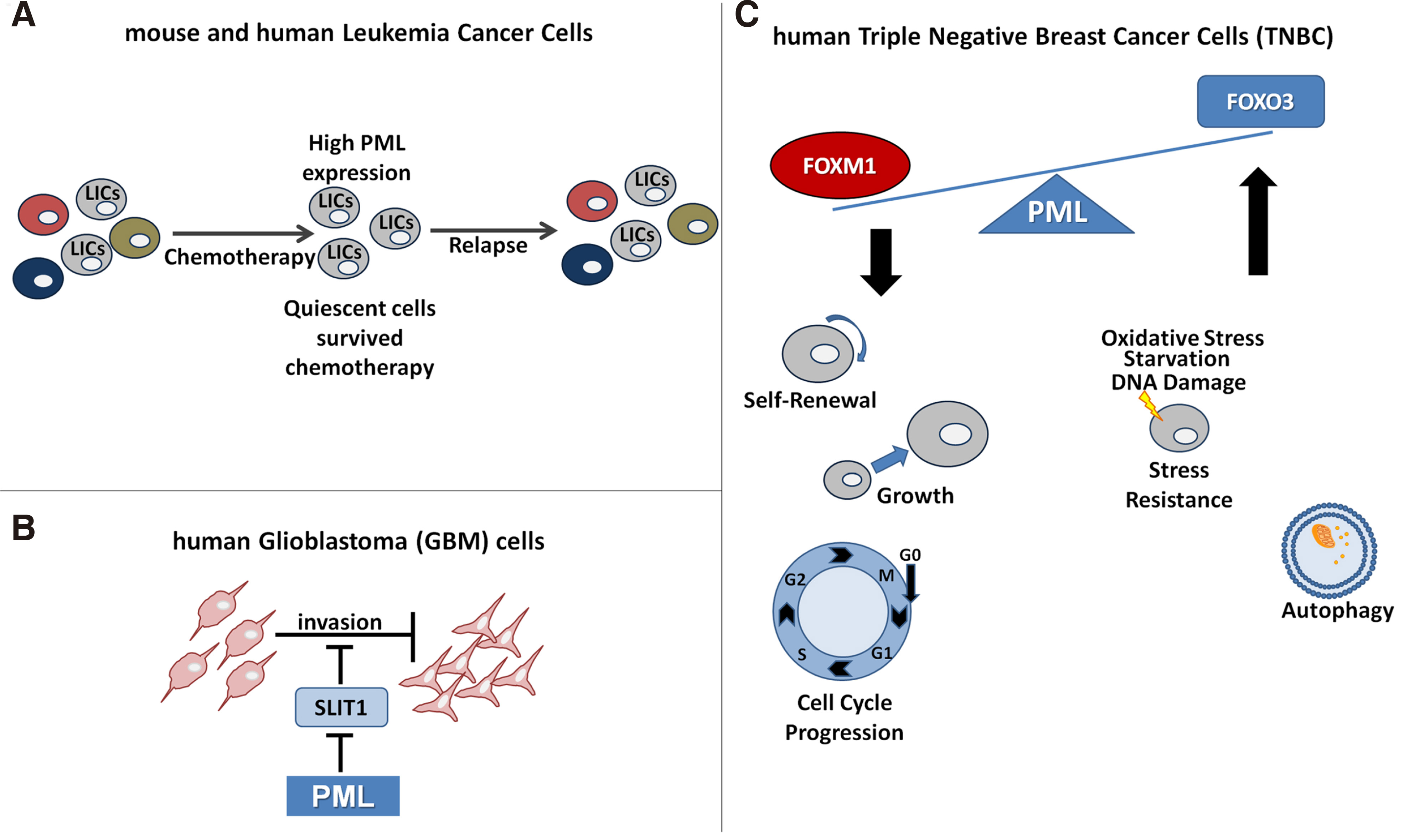
The above studies support a tumor suppressor role for PML. However, other studies revealed that in a cell context-dependent way, PML also shows pro-oncogenic properties, contributing to tumor cell metabolism and self-renewal capacity. Indeed, PML appears to be elevated in several cases of leukemia and more specifically in chronic myeloid leukemia (CML). In fact, patients with CML and low PML expression had an overall better response to treatment, an observation that contrasts with a tumor suppressor role in solid tumors. Deletion of PML reduced the population of leukemia initiating cells (LICs), confirming a role in the maintenance of normal and leukemia stem cells (Ito et al. 2008) (Fig. 2A). A pro-survival role for PML was found in the established breast epithelial MCF10A cell line. By activating FAO, PML was able to protect MCF10A cells from anoikis and support luminal cell filling in a 3D culture model (Carracedo et al. 2012). In a study using liver CSCs, arsenic trioxide-mediated PML degradation led to the repression of Oct4 gene expression and the reduction of CSC population (Tang et al. 2016). Finally, in human glioblastoma cells, PML shows a pro-migratory activity via repression of Slit1, a critical regulator of axon guidance (Amodeo et al. 2017) (Fig. 2B).
Fig. 2. Multiplicity of PML effects in a cell-context dependent manner in cancer.
(A) PML contributes to the maintenance and chemotherapy resistance of leukemia initiating cells (LICs). (B) It acts as pro-migratory factor in glioblastoma cells via EZH2-mediated epigenetic suppression of SLIT1 expression. (C) PMLIV ectopic expression suppresses proliferation and self-renewal ability in TNBC cells and enhances cell stress resistance through repression of FOXM1 and modulation of FOXO3 protein respectively.
High PML RNA levels correlate with poor prognosis in mesenchymal type Glioblastoma and triple negative breast cancer (TNBC) tumor subsets (Amodeo et al. 2017; Ponente et al. 2017). Interestingly, these effects are context specific, as they are not observed in other tumor types or intrinsic subtypes. It has also been demonstrated that PML cooperates with HIF1A to enhance the expression of several genes implicated in cell migration and invasion in TNBC cells (Martín-Martín et al. 2016; Ponente et al. 2017), although earlier studies found that PML strongly inhibits the mTOR-HIF-angiogenic response in a non-cancer mouse model (Bernardi and Pandolfi, 2007).
In a recent publication, we showed that the PMLIV isoform, known to promote apoptosis and senescence upon DNA damage (Nisole et al. 2013), strongly inhibits proliferation and stem cell enriched sphere formation (Sachini et al. 2019) of breast MDA-MB-231 and Glioma U87MG cells (Tampakaki et al. 2021). PMLIV, but no other isoforms, such as PML I or III, were found to antagonize pro-proliferative signaling of Forkhead Box M1 (FOXM1) (via both protein binding and RNA reduction) while de-repressing Forkhead Box O3 (FOXO3) functions, associated with cell survival and resistance to cellular stress (Sachini et al. 2019) (Fig. 2C). Interestingly, other studies found that the PMLI isoform had tumor-promoting roles in the above systems (Alhazmi et al. 2020), thus isoform specific functions should also be explored. The aforementioned studies, mostly based on PML ablation or ectopic expression, resulted in heterogeneous results that may be due to the diverse genetic and phenotypic background of the cells under study, the differential expression and role of the various PML isoforms, or both.
Cytoplasmic PML (cPML) -specific biological functions are also described, namely the enhancement of TGF signaling leading to growth arrest and cellular senescence (Lin et al. 2004) or facilitation of cell migration and EMT (Buczek et al. 2016). Most importantly, cPML associated with mitochondrial membranes was reported to behave as a tumor suppressor by regulating calcium flux and inhibiting autophagy in a p53 dependent manner (Missiroli et al. 2016).
Taken together, the above results reveal a complex relationship between PML and cancer. Different isoforms of PML may function as either tumor suppressors or tumor promoters, depending on the cell type, genetic context and interacting factors (Datta et al. 2021).
Tissue Specific Stem Cells
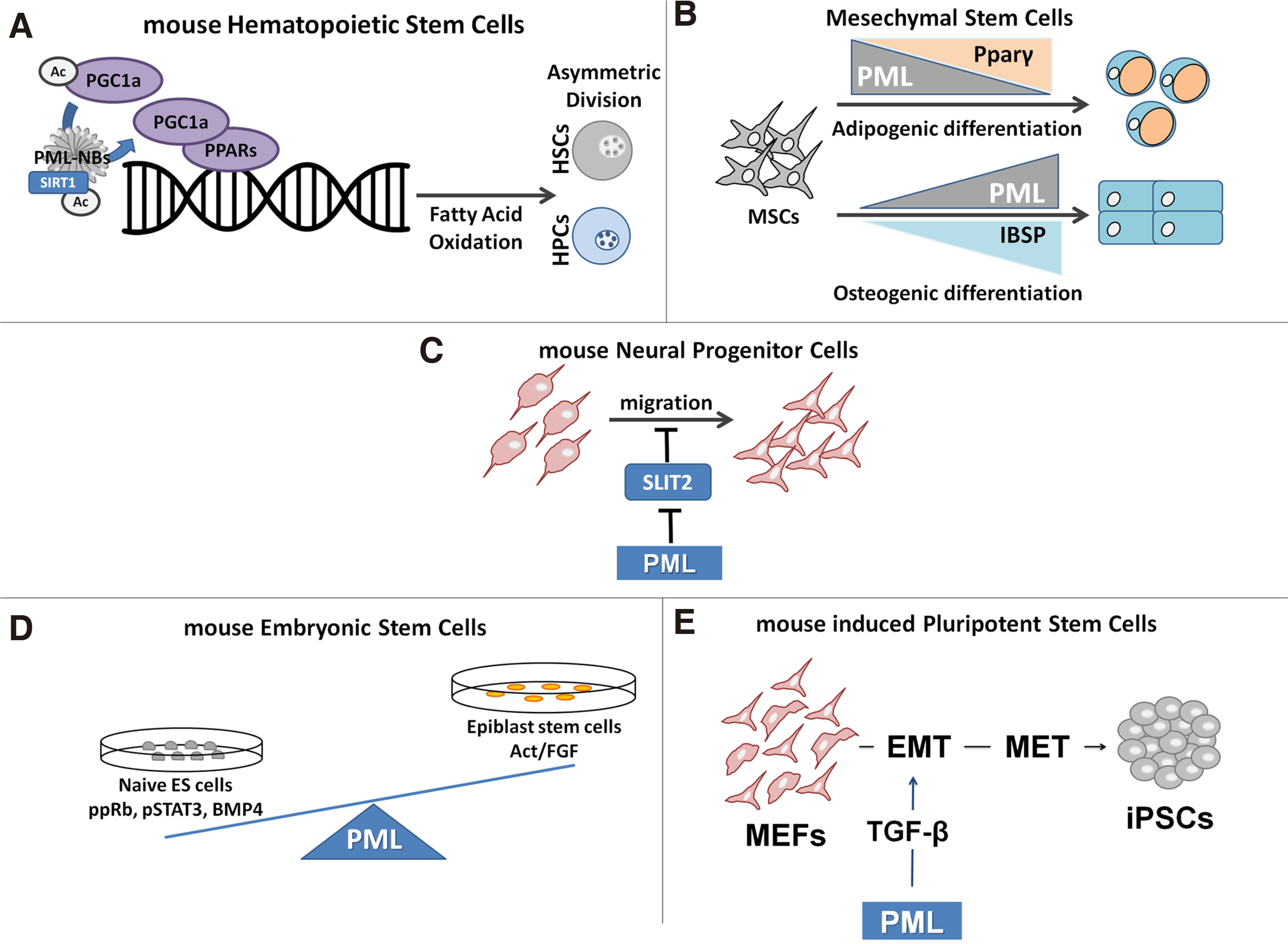
Promyelocytic leukemia protein is important for mouse hematopoietic stem cell (HSC) self-renewal. It regulates the growth and proliferation of HSCs via the phosphatidylinositol 3-kinase (PI3K)/Akt pathway. In this cell-type, PML regulates the asymmetric division and maintenance of HSCs via its involvement in the deacetylation of the PPARγ coactivator 1A (PGC1A) by SIRT1, which leads to activation of the PPAR pathway and fatty acid oxidation (FAO). Fatty acid oxidation, in turn, stimulates the asymmetric cell divisions of HSCs, which allows their self-renewal. Pharmacological inhibition of FAO by etomoxir favors symmetric divisions and exhaustion of HSCs. Deletion of PPARδ or PML, as well as inhibition of FAO, results in the differentiation of HSCs, while activation of PPARδ increases asymmetric cell division, which suggests that the PML-PPARδ-FAO axis regulates the choice between self-renewal and differentiation in HSCs (Ito et al. 2008; Ito et al. 2012; Lallemand-Breitenbach and de Thé, 2012) (Fig. 3A). Promyelocytic leukemia protein is also required for cell lineage determination in mammary gland progenitor cells (Li et al. 2009). The role of PML in Mesenchymal Stem Cells (MSC) is not clear. PML-/- mice had increased adipose cell number under high fat diet when compared with control individuals. Moreover, PML was shown to inhibit adipogenic differentiation of 3T3-L1 mouse cells (Kim et al. 2011). In a more recent study, Morganti et al found that PML is essential for efficient adipogenic differentiation of mouse MSCs through modulation of autophagy (Morganti et al. 2019). In another report, PML was acting as inhibitor of cell proliferation and promoter of osteogenic differentiation in human bone marrow mesenchymal stem cells via upregulation of bone integrin-binding sialoprotein (IBSP, bone sialoprotein) (Sun et al. 2013) (Fig. 3B).
Fig. 3. The Roles of PML and PML NB in stem cells.
(A) PML is required for HSC pool maintenance by promoting asymmetric divisions via the PPAR- -FAO pathway. (B) PML is essential for mMSC differentiation to occur in the initial steps of adipogenesis. PML is induced and promotes hMSC osteogenic differentiation. (C) PML contributes to NPC migration by inhibiting SLIT2 expression. (D) PML is required for ESC self-renewal and naïve pluripotent state by regulating cell cycle progression, transcription factor expression and signaling pathways activities. (E) PML potentiates the reprogramming of MEFs into pluripotency via regulation of the sequential EMT and MET required in the early steps.
PML has important functions in the nervous system. In the developing mouse neocortex, PML is expressed only in Neural Progenitor Cells (NPC), where it seems to limit their proliferation, via pRB regulation, in favor of differentiation. By disrupting the balance between proliferation and differentiation, PML loss in mice leads to a thinner cortex wall (Regad et al. 2009). In adult neurogenesis, PML functions as a positive regulator of NPC migration via repression of SLIT2 gene expression, an anti-migratory factor (Amodeo et al. 2017) (Fig. 3C).
Pluripotent stem cells
Embryonic stem cells derived from early embryos and induced Pluripotent Stem Cells (iPSCs) are characterized by the ability to self-renew indefinitely and to differentiate into all the three germ lineages—ectoderm, mesoderm, and endoderm—of the developing embryo. Their pluripotency is governed by a core network of master regulatory transcription factors including pluripotent Oct4, Sox2 and Nanog, extrinsic signaling pathways and epigenetic complexes (Jaenisch and Young, 2008).
We described a novel role for PML and PML nuclear bodies in naïve and induced pluripotency (Hadjimichael et al. 2017). Promyelocytic leukemia protein promotes ESC cycle progression by maintaining the inactive (phosphorylated) form of Rb and the active form of Stat3. In the absence of PML, the G1 phase of ESCs is prolonged at the expense of S phase. Furthermore, PML-deficient ESCs exhibit morphological, transcriptomic and metabolic properties distant from the naive and closer to the primed pluripotent state. Naïve pluripotency factors (Nanog, Dazl, Cadherin1) and pathways (LIF/BMP) are downregulated, whereas primed state markers (Otx2, Fgf5) and activin pathway factors are upregulated (Fig. 3D). Accordingly, PML is required for the reprogramming of mouse embryonic fibroblasts (MEFs) to pluripotency via a TGF-β dependent early EMT and MET transition (Fig. 3E). In addition to its effect on the ESCs state, we have also shown that PML is involved in cell lineage decision making, as PML is required for the endodermal specification, while in its absence (PML-/-), the mesodermal lineage is favored (Hadjimichael et al. 2017).
In another recent report, a role for PML bodies in ESC 3D nuclear organization was described. Binding of PML bodies to 300 kb of the short arm of the Y chromosome was required to inhibit methylation by DNMT3a and transcriptional repression of neighboring Y linked genes (Kurihara et al. 2020) (Fig. 4C). In addition, PML ablation was reported to direct ESC towards the two-cell totipotent state. The reason for the difference between this and the aforementioned publication (Hadjimichael et al. 2017; Kurihara et al. 2020) on ESC state changes induced by PML loss remains unclear. It might be due to cell type, cultivation conditions and PML ablation method differences. Interestingly, it is worth noting that both publications conclude that PML safeguards naïve ESC pluripotency.
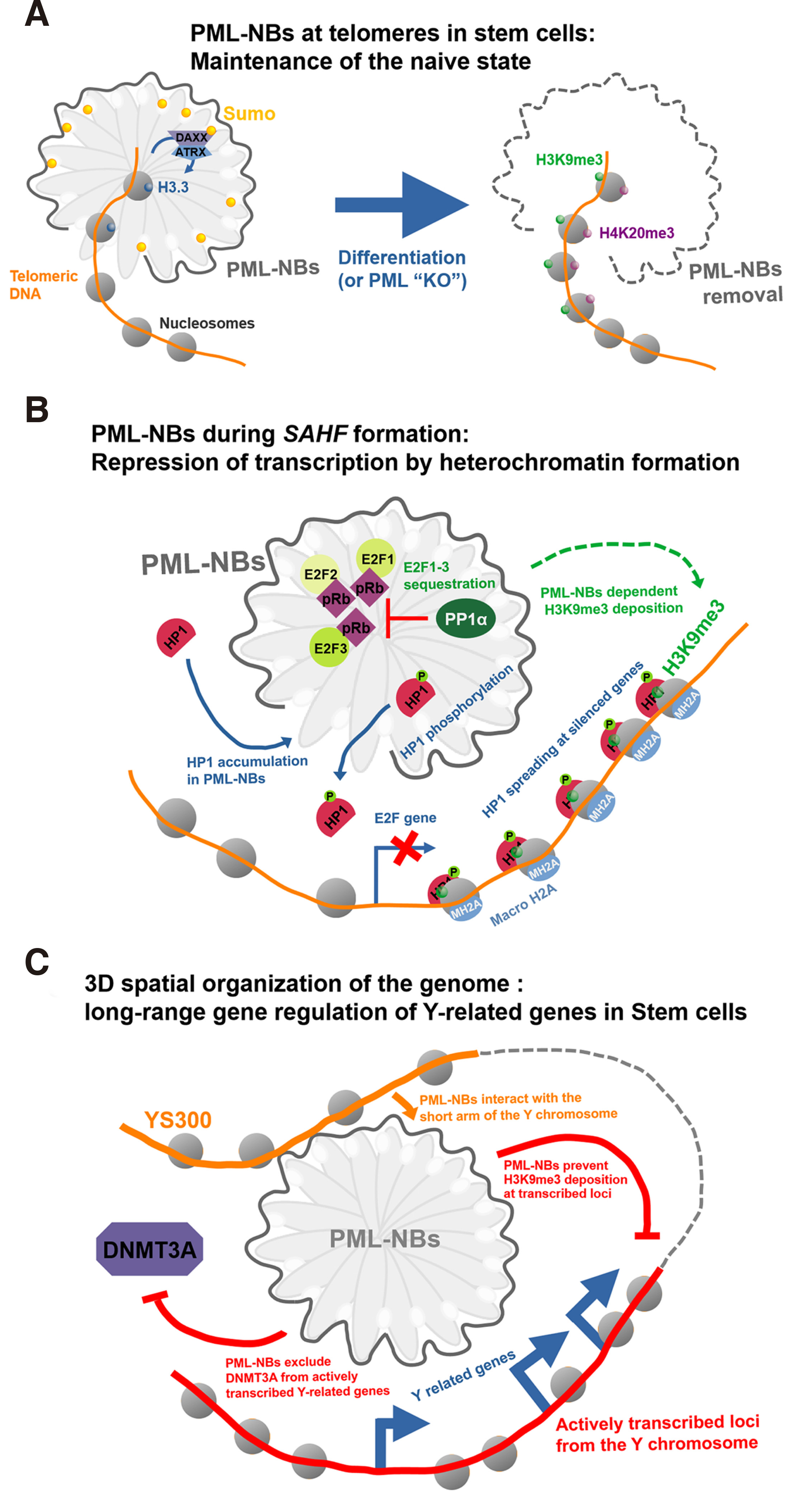
Fig. 4. Examples of PML-NBs contributions in regulating chromatin dynamics.
(A) PML-NBs regulate chromatin dynamics at telomeres in ESC. In the undifferentiated ESC state, telomeric chromatin is enriched in histone variant H3.3 in a PML-NB dependent fashion, whereas upon differentiation (or experimental PML depletion), the local chromatin adopts a heterochromatin state enriched with H3K9me3 and H4K20me3. (B) PML-NBs in senescence-associated heterochromatin foci (SAFHs) formation. PML-NBs contribute to E2F-dependent genes silencing by sequestration of E2F factors and deposition of H3K9me3 and HP1 and macro-H2A spreading. (C) Spatial re-arrangement of chromosomes and long-range gene regulation mediated by PML-NBs in ESC. PML-NBs interaction with the short arm of the Y chromosome (YS300) maintains the expression of several Y related genes via exclusion of DNMT3A and prevention of H3K9me3 marks addition.
The functions of PML in ESC reveal a pro-oncogenic potential similar to that already mentioned, and place PML in the category of tumor suppressors acting as double agents (Datta et al. 2021).
PML and Epithelial-Mesenchymal transitions
Epithelial mesenchymal transition (EMT), and its reverse, MET, are two cell-fate processes, known to be critical for cancer metastases (Jolly et al. 2017). In this context, we have shown that PML is required for the reprogramming of fibroblasts back to the pluripotent state through a mechanism involving a TGF-β dependent early EMT-MET transition (Hadjimichael et al. 2017).
Epithelial mesenchymal transition is a process that is normally involved in various developmental stages but also in the wound healing process. During malignant progression, epithelial cells undergo an EMT like switch by losing apical-basal polarity and tight junctions, a process important for cell migration and metastasis (Bill and Christofori, 2015). Upon reaching their target organ, these migrating cells follow MET that allows metastatic growth (Tsai et al. 2012). Epithelial mesenchymal transition can be induced by several cellular pathways such as growth factors, TGF-β, WNT, and Notch signaling. As noted above, the cytoplasmic isoform of PML (PMLVII) induces TGF-β signaling by SMAD2 and SMAD3 phosphorylation in mouse embryonic fibroblasts (MEFs) (Lin et al. 2004) and promotes EMT in prostate cancer cells via TGF-β/SMAD signaling, enabling cancer cells to acquire a more aggressive and invasive phenotype (Buczek et al. 2016). Conversely, in the absence of the EMT factors Snail and TGF-β, the Inhibitor of DNA Binding 1 (ID1) factor can induce MET by antagonizing TWIST1 activity (Stankic et al. 2013). Interestingly, ID1 was earlier identified as a member of a group of genes that enables lung cancer colonization (Gupta et al. 2007). Along with the ID1 role in HIF stabilization and angiogenesis (Kim et al. 2007), IDs emerge as key factors in both the early (angiogenic) and late (colonization metastatic) process. In line with those studies, a recent work by our group (A. P. Vogiatzoglou et al, unpublished) demonstrates that knocking down PML results in higher lung metastasis in xenografted mice that correlates with high ID1 levels, especially in lung–derived tumor cells. These results point to divergent effects of PML and its isoforms in EMT and cell migration/metastasis, mediated by multiple pathways and stages of the metastatic process that involve EMT and ID factors, migration inhibitors and angiogenesis. Thus, once again, the final PML effect is highly complex and context dependent.
PML and Heterochromatin
Though there is no evidence that PML-NBs act as direct regulators of transcription, many PML-NBs clients are chromatin modifying proteins, raising interest in PNL-NBs as chromatin content regulators and, more generally, as indirect regulators of gene expression. This clients list includes, most notably: histone acetyltransferases TIP60, MOZ and CREB-Binding Protein CBP, histone deacetylase SIRT1 and HDAC7, histone 3 lysine 9 methyltransferase SETDB1 (all involved in transcription regulation); HP1, DEK and DAXX (death associated protein), involved in transcription repression by heterochromatin formation; the HIRA histone chaperone complex (formed by HIRA, ASF1A, CABIN-1 and UBN1 proteins) and DNA demethylation enzymes such as DNMT3A and TET2 (LaMorte et al. 1998; Lehming et al. 1998; von Mikecz et al. 2000; Wang et al. 2004; Corpet et al. 2020;)
While the direct interaction of PML-NBs with chromatin has been the subject of controversy, it is well established that they are essential to the control of chromatin quality content in silenced heterochromatic loci telomeres and peri-centromeric regions (Luciani et al. 2005, 2006; Oh et al. 2009; Corpet et al. 2014;). Telomeres are essential to protect chromosomes from degradation by shortening or fusion with other chromosomes during cell proliferation. Maintenance of Telomeres integrity is therefore fundamental to the proliferating capacity of human stem cells (Liu, 2017). This maintenance is ensured by the telomerase proteins, which directly interact with the telomeric region and, as such, require DNA access through chromatin (Blackburn, 2000). The epigenetic marks found at Telomeres are a source of controversy (Udroiu and Sgura, 2020), and telomeres from mammalian cells have been both found featuring canonical heterochromatin marks (H3K9me2, H3K9me3 and H4K20me3) and euchromatin ones (such as H3K27ac).
In contrast to somatic cells, mouse ESC and iPSC feature a lower abundance of heterochromatin marks at telomeres. However, upon differentiation, descendants of both cell types have been reported to present an increasing level of H3K9me3 and H4K20me3 at telomeres, indicating the existence of a mechanism dynamically regulating chromatin marks pattern during cell-fate changes (Udroiu and Sgura, 2020) (Fig. 4A). Telomeres in mESCs are also enriched with the alpha-thalassemia mental retardation syndrome, X-linked ATRX protein and the histone variant H3.3 (Wong et al. 2010; Chang et al. 2013). ATRX is an essential chromatin remodeling protein that bears a SWI/SNF helicase/ATPase domain. It also interacts with death-associated protein DAXX, a histone chaperone important for H3.3 variant incorporation and involved in apoptosis (Dyer et al. 2017). Loss of function of ATRX or DAXX decreases H3.3 marks at telomeres, which induces cell senescence (Chang et al. 2013). Promyelocytic leukemia protein–nuclear bodies orchestrate H3.3 incorporation at telomeres by constitutively recruiting DAXX through sumoylation of PML B-BOX1 domain (Zhu et al. 2005; Percherancier et al. 2009). Correspondingly, deletion of PML results in a loss of ATRX recruitment at telomeres and, consequently, reduced local H3.3 incorporation, indicating PML’s involvement in sustaining epigenetic stability at telomeres in mESCs (Chang et al. 2013).
It is worth noting that PML has also been implicated as contributing to peri-centromeric heterochromatin regulation. PML-NBs have been found to be associated with the human peri-centromeric region after replication of the DNA during the cell cycle, which would suggest a role in re-condensation of DNA and establishment of heterochromatin at these sites (Luciani et al. 2006). However, these regions did not feature heterochromatin methylation marks H3K9me3 and H4K2me3, but rather the H3.3 histone variant (Drané et al. 2010; Goldberg et al. 2010). While components of the H3.3 assembly pathway (ATRX, DAXX) were co-localised at PML-NBs at peri-centrosomes in these cells, it is still unclear whether their assembly occurred in a PML-NBs dependent fashion (Corpet et al. 2014). A study performed in MEFs even demonstrated that PML-NBs could be an antagonist factor on the H3.3 deposition pathway, as depletion of PML resulted in an increase in H3.3 incorporation at the peri-centromeric repeats (Shastrula et al. 2019). Further studies are seemingly required in order to fully apprehend PML-NBs contribution to heterochromatin formation at peri-centrosomes. This is increasingly important as mis-expression of peri-centrosomal region genes and more generally centrosomes amplifications, are features of cancer (Gordon et al. 2012; Singh et al. 2020; Mittal et al. 2021).
PML-NB regulation of heterochromatic regions is not restricted to somatic maintenance of heterochromatin but is also involved in cancer response triggered senescence (Wang et al. 1998; Trotman et al. 2006). Senescent cells are characterized by a permanent cell-cycle arrest that can occur following various stresses internal or external to the cell, such as telomeres attrition or oncogenic factors (Hayflick and Moorhead, 1961). Senescent cells are characterized by repression of cell-cycle related genes, such as those regulated by E2F1-3 factors and by expression of apoptosis genes. PML-NBs contribute to the anti-tumoral response by inducing and sustaining senescence (Ferbeyre et al. 2000) through different pathways. PML-NBs were originally found in increased size and number in senescent cells. Furthermore, overexpression of PML induces early senescence entry, while PML-/- mutants are unresponsive to senescence induction by oncogenic factor RAS (Ferbeyre et al. 2000; Pearson et al. 2000; Bischof et al. 2002). During senescence, each chromosome condenses into senescence associated heterochromatin foci (SAHF) that feature enrichment of H3K9me3 mark, macroH2A histone variant and recruitment of HP1 heterochromatin factor, together leading to effective silencing of replication and proliferation related genes (Corpet and Stucki, 2014). This genome-wide chromatin remodeling requires PML-NBs that, in response to p53/pRb, recruit and sequester components of the HIRA complex, including the histone chaperon ASF1A, and E2F1-3 factors (Zhang et al. 2005, 2007; Takahashi et al. 2007). While it is unclear why the HIRA complex is sequestered at PML-NBs, as no H3.3 variant are found at SAHF, it is well established that this re-location is required for SAHF formation (de Stanchina et al. 2004; Corpet et al. 2014). Similarly, E2Fs accumulation at PML-NBs renders them unavailable for gene activation and eventually forestall re-entry into the cell cycle. Finally, HP1 accumulation at PML-NBs occurs following p53/pRB senescence induction and precedes SAHF formation (Fig. 4B). Indeed, HP1 at heterochromatin is complexed with H3K9me3 marked chromatin, and HP1 spreading can then occur via oligomerization. The local increase in HP1 concentration by PML-NBs recruitment may help to pass the concentration threshold for phase separation that will render soluble heterochromatin into a phase-separated droplet state that will limit access of the DNA template to the transcription pioneer factors and promote gene silencing. Defects in the re-localization of HIRA complex or HP1 at PML-NBs lead to SAHF formation failure (Zhang et al. 2005; Banani et al. 2016). While they do not contain SAHF, PML-NBs in senescent cells do associate with promoters of E2F-regulated genes, which ensure their silencing by H3K9me3 and HP1 spreading (Talluri and Dick, 2014). Retinoblastoma protein (pRb) is required for complete senescence commitment via PML-dependent heterochromatin SAHF formation in response to RAS, as Rb mutants unable to interact with PML (LXCXE domain RB mutants) do not fully establish heterochromatin at E2Fs genes (Ferbeyre et al. 2000; Vernier et al. 2011). Further investigation will be necessary to fully understand PML contribution to heterochromatin formation during senescence, and in particular its involvement in establishing the H3K9me3 mark.
Finally PML-NBs are reported to induce latent/quiescent HSV-1 genomes chromatinization through a PML NB/Histone H3.3/ H3.3 Chaperone Axis. This mechanism accounts for the well-known anti-viral action of PML (Cohen et al. 2018).
PML and regulation of active transcription
Promyelocytic leukemia protein contribution to epigenetic marks reshaping during stem cell differentiation is not limited to telomeres silencing but extends to the core body of chromosomes and is essential to the initiation and commitment of cell-fate decision making (Gaspar-Maia et al. 2011). Promyelocytic leukemia PML-NBs have been found to be associated at actively transcribed loci but do not incorporate nascent RNA, suggesting a role in transcription regulation and not transcription per se (Boisvert et al. 2000; Boisvert et al. 2001; Wang et al. 2004).
In the past, proximity mechanisms that alter chromatin configuration in the vicinity of the PML-NBs were shown to regulate gene expression. PML-NBs and Satb1 were shown to contact the major histocompatibility complex (MHC) class I locus and enhance active transcription (Kumar et al. 2007). In this vein, we have shown that the Interferon induced MHC class II locus delocalizes close to PML NB upon previous treatment with interferon IFN-γ. While in this state, the DRA locus is poised by high levels of H3K4me2 marks mediated by mixed-lineage leukemia (MLL) factor and features an open chromatin state, all of which favors subsequent transcription re-activation. PML-NB contribution to 3D spatial chromosome re-arrangement is therefore able to set-up gene transcription and maintain a memory of past gene activation (Gialitakis et al. 2010).
Part of the difficulty of understanding the contribution of PML-NBs to gene regulation resides in the fact that while nucleoplasmic PML are soluble and therefore can be subjected to ChIP, PML-NBs, which are insoluble and form liquid-liquid phase separation aggregates, cannot. Using a novel approach they engineered (Alap-seq) that relies on sequencing DNA fragments retrieved from chromatin biotinylated by an APEX peroxidase fused to PML-NBs, the Miyanari lab circumvented the short-coming of chromatin immunoprecipitation of insoluble PML-bodies and uncovered new genome-wide association of PML-NBs in mESCs (Kurihara et al. 2020). Their data indicated that most PML-NBs were associated with core promoters and transcription enhancers of known regulatory regions, such as telomeres. Interestingly, these PML-NB associated core-promoter regions featured marks of active transcription elements, with open chromatin state, enrichment of H3K27ac and H3K4me3 active marks and depletion of H3K9me3 and H3K27me3 repressive marks. These hot-spot PML peaks were poorly matched when using a PML ring mutant unable to form bodies, indicating that the data obtained was clearly mirroring chromatin association of PML-NBs. Among these new PML-NBs hot-spots was YS300, the short arm of the Y chromosome. Consequently, deletion of PML in mESCs was found to mis-regulate Y-linked genes (Kurihara et al. 2020). PML-NB recruitment at YS300 promotes Y-linked genes expression by exclusion of DNMT3A (Fig. 4C). PML-NB liquid-liquid phase separation properties may be responsible for this exclusion, generating a close environment prone to gathering transcription factors as detailed in the recent transcription activation model by the Young group (Boija et al. 2018). It is also possible that this specific exclusion of DNMT3A, but not DNMT3B, is achieved by a post-translational modification of DNMT3A, or one of its partners, which could in turn be mediated by the PML-NBs. Post-translational modification is known to condition access to liquid-liquid phase separated environment (Schuster et al. 2018). Finally, an additional mechanism that may antagonize gene suppression by DNA methylation involves TET2 binding and recruitment of PML to methylated promoters (Song et al. 2018).
While the above data supports a more direct role of PML-NBs in gene regulation, it is still unclear how PML-NBs render either a transcriptionally repressive or active chromatin state in relation to the genomic positions of their target. Sequence motif analysis of PML-NBs hot-spots obtained by Alap-seq did not reveal any specific DNA motifs. One possibility would be that monomeric PML of PML-NBs are locally recruited by specific transcription or chromatin bound factors at loci where they generate a liquid-liquid phase separation environment. This recruitment may involve post-translational modifications, such as sumoylation of the telomeric SIM proteins TRF1 and TRF2, which allows recruitment of PML at telomeres (Potts and Yu, 2007). PML-NBs themselves undergo sumoylation, and recruit SUMO1, so it may be possible to recruit additional factors at active core-promoters through their sumoylation (Ivanschitz et al. 2013).
Conclusions
In the last decade, in addition to the classical tumor suppressor function, recent publications have enriched the PML repertoire with new processes, such as regulation of cell migration, tumor cell survival, autophagy, stem cell self-renewal and chromatin modifying activities. In contrast to its initially recognized role as tumor suppressor, these reports expand its functional diversity and assign pro-oncogenic potential to PML. Increasing recent evidence connects these processes to stem cell physiology. Studies of PML in leukemia showed its involvement in maintaining HSC self-renewal via PPAR-FAO signaling. In embryonic or adult neural progenitor cells, PML potentiates their differentiation and migration, respectively. Other studies have pointed to a new role for PML in pluripotent stem cell fate decisions. In mouse ESC, PML is required for their self-renewal and the maintenance of the naïve state, and potentiates the somatic cell reprogramming into pluripotency. Taken together, these studies suggest a common role of PML in stem cells of diverse origins.
The complexity of the PML landscape, its many isoforms and multiple clients, underlies its functional diversity and its frequently contradictory effects in different contexts. Deciphering the PML–code, which will not only interpret but also predict its biological response in different set ups, is a challenging task. To this end, a number of questions are open to investigation:
1. Since the mechanisms or distinct gene targets that determine the type of tumor response by PML are not clearly understood, there is a need for studies in tumor models that faithfully recapitulate specific human cancer types and their oncogenic driver contexts in combination with genome- and proteome-wide analysis. This will permit reliable predictions of the role of PML or its targets in distinct cancer settings.
2. Knowledge of the role of structure –function diversity of PML protein isoforms and their variable stoichiometry in distinct cell contexts is required in order to understand the overall PML impact.
3. Promyelocytic leukemia protein is a “double TSG agent”. Strikingly, many PML-NB “clients” are also such factors. This begs the question as to whether the PML duality stems from the preferential or dominant interaction(s) with its oncogenic or tumor suppressor clients in an isoform and/or cell specific way.
4. The elucidation of PML functions in cell fate decisions is far from complete. In this very promising field, the role of PML in stress response and cell regeneration might offer clinical applications.
5. Promyelocytic leukemia protein has been involved in both establishment of heterochromatin and activation of gene expression. The determinants of such specificities remain unclear.
Overall, thorough studies designed to integrate cell type, genomic and proteomic data along with informatics analysis are necessary to evaluate the PML effect in different set-ups, along with the prevailing cancer or signaling pathway involved.
Acknowledgements
The authors are grateful for the technical support of T. Makatounakis and G. Vrentzos.
Declarations
Author contributions
A.P.V., F.M., M.M., J.P., A.K. wrote, edited, and revised the original drafts. A.P.V. and F.M. prepared the figures. All authors have read and agreed to the published version of the manuscript.
Funding
This research is co-financed by Greece and the European Union (European Social Fund- ESF) through the Operational Programme «Human Resources Development, Education and Lifelong Learning 2014-2020» in the context of the project “The role of Promyelocytic Leukemia Protein in normal and cancer stem cells” (MIS 5048184).
Abbreviations
APL, acute promyelocytic leukemia ; CML, chronic myeloid leukemia ; c-Myc, MYC Proto-Oncogene ; CSCs, cancer stem cells ; DNMT, DNA methyltransferase ; EMT, Epithelial-Mesenchymal Transition ; FAO, fatty acid oxidation ; FOXM1, Forkhead Box M1 ; FOXO3, Forkhead Box O3 ; HIF, Hypoxia Inducible Factor ; HIF1A, Hypoxia Inducible Factor 1 Subunit Alpha ; HP1, Heterochromatin Protein 1 ; HSCs, hematopoietic stem cells ; ID1, Inhibitor of DNA Binding 1 ; iPSCs, induced Pluripotent Stem Cells ; Klf4, Kruppel Like Factor 4 ; LICs, leukemia initiating cells ; MCF10A, breast epithelial cell line ; mESCs, mouse embryonic stem cells ; MHC, major histocompatibility complex ; MLL, mixed-lineage leukemia ; MSCs, Mesenchymal Stem Cells ; mTOR, mechanistic target of rapamycin ; Oct4, Octamer-Binding Protein 4 ; PML, Promyelocytic leukemia protein ; PML-NBs, Promyelocytic leukemia protein–nuclear bodies ; PPAR, Peroxisome Proliferator Activated Receptor ; pRB, retinoblastoma protein/RB Transcriptional Corepressor 1RAR, retinoic acid receptor ; RBCC, RING finger, B-box, and coiled-coil ; SAHF, senescence associated heterochromatin foci ; Sox2, SRY-Box Transcription Factor 2 ; TET2, Ten-Eleven Translocation 2 ; TGF-β, Transforming Growth Factor Beta ; TME, tumor microenvironment ; TME, tumor microenvironment ; TNBC, triple negative breast cancer cell line ; TRIM, tripartite motif family ;References
Alhazmi N., Pai C.P., Albaqami A., Wang H., Zhao X., Chen M., Hu P., Guo S., Starost K., Hajihassani O., Miyagi M., Kao H.Y. (2020). The promyelocytic leukemia protein isoform PML1 is an oncoprotein and a direct target of the antioxidant sulforaphane (SFN). Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1867: 118707.
Amodeo V., A D., Betts J., Bartesaghi S., Zhang Y., Richard-Londt A., Ellis M., Roshani R., Vouri M., Galavotti S., Oberndorfer S., Leite A. P., Mackay A., Lampada A., Stratford E. W., Li N., Dinsdale D., Grimwade D., Jones C., Nicotera P., Michod D., Brandner S., Salomoni P. (2017). A PML/Slit Axis Controls Physiological Cell Migration and Cancer Invasion in the CNS. Cell Reports 20: 411-426.
Banani S. F., Rice A. M., Peeples W. B., Lin Y., Jain S., Parker R., Rosen M. K. (2016). Compositional Control of Phase-Separated Cellular Bodies. Cell 166: 651-663.
Beck B., Blanpain C. (2013). Unravelling cancer stem cell potential. Nature Reviews Cancer 13: 727-738.
Bernardi R., Guernah I., Jin D., Grisendi S., Alimonti A., Teruya-Feldstein J., Cordon-Cardo C., Celeste Simon M., Rafii S., Pandolfi P. P. (2006). PML inhibits HIF-1α translation and neoangiogenesis through repression of mTOR. Nature 442: 779-785.
Bernardi R., Pandolfi P. P. (2007). Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nature Reviews Molecular Cell Biology 8: 1006-1016.
Bill R., Christofori G. (2015). The relevance of EMT in breast cancer metastasis: Correlation or causality?. FEBS Letters 589: 1577-1587.
Bischof O. (2002). Deconstructing PML-induced premature senescence. The EMBO Journal 21: 3358-3369.
Blackburn E. H., (2000). The end of the (DNA) line. Nature Structural Biology 7: 847-850.
Boija A., Klein I. A., Sabari B. R., Dall’Agnese A., Coffey E. L., Zamudio A. V., Li C. H., Shrinivas K., Manteiga J. C., Hannett N. M., Abraham B. J., Afeyan L. K., Guo Y. E., Rimel J. K., Fant C. B., Schuijers J., Lee T. I., Taatjes D. J., Young R. A. (2018). Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 175: 1842-1855.e16.
Boisvert F.M., Hendzel M. J., Bazett-Jones D. P. (2000). Promyelocytic Leukemia (Pml) Nuclear Bodies Are Protein Structures That Do Not Accumulate RNA. Journal of Cell Biology 148: 283-292.
Boisvert F.M., Kruhlak M. J., Box A. K., Hendzel M. J., Bazett-Jones D. P. (2001). The Transcription Coactivator Cbp Is a Dynamic Component of the Promyelocytic Leukemia Nuclear Body. Journal of Cell Biology 152: 1099-1106.
Borden K. L.B. (2008). Pondering the puzzle of PML (promyelocytic leukemia) nuclear bodies: Can we fit the pieces together using an RNA regulon?. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1783: 2145-2154.
Buczek M. E., Miles A. K., Green W., Johnson C., Boocock D. J., Pockley A. G., Rees R. C., Hulman G., van Schalkwyk G., Parkinson R., Hulman J., Powe D. G., Regad T. (2016). Cytoplasmic PML promotes TGF-β-associated epithelial–mesenchymal transition and invasion in prostate cancer. Oncogene 35: 3465-3475.
Carnero A., Lleonart M. (2016). The hypoxic microenvironment: A determinant of cancer stem cell evolution. Inside the Cell 1: 96-105.
Carracedo A., Weiss D., Leliaert A. K., Bhasin M., de Boer V. C.J., Laurent G., Adams A. C., Sundvall M., Song S. J., Ito K., Finley L. S., Egia A., Libermann T., Gerhart-Hines Z., Puigserver P., Haigis M. C., Maratos-Flier E., Richardson A. L., Schafer Z. T., Pandolfi P. P. (2012). A metabolic prosurvival role for PML in breast cancer. Journal of Clinical Investigation 122: 3088-3100.
Chang F. T. M., McGhie J. D., Chan F. L., Tang M. C., Anderson M. A., Mann J. R., Andy Choo K. H., Wong L. H. (2013). PML bodies provide an important platform for the maintenance of telomeric chromatin integrity in embryonic stem cells. Nucleic Acids Research 41: 4447-4458.
Cohen C., Corpet A., Roubille S., Maroui M. A., Poccardi N., Rousseau A., Kleijwegt C., Binda O., Texier P., Sawtell N., Labetoulle M., Lomonte P. (2018). Promyelocytic leukemia (PML) nuclear bodies (NBs) induce latent/quiescent HSV-1 genomes chromatinization through a PML NB/Histone H3.3/H3.3 Chaperone Axis. PLOS Pathogens 14: e1007313.
Corpet A., Kleijwegt C., Roubille S., Juillard F., Jacquet K., Texier P., Lomonte P. (2020). PML nuclear bodies and chromatin dynamics: catch me if you can!. Nucleic Acids Research 48: 11890-11912.
Corpet A., Olbrich T., Gwerder M., Fink D., Stucki M. (2014). Dynamics of histone H3.3 deposition in proliferating and senescent cells reveals a DAXX-dependent targeting to PML-NBs important for pericentromeric heterochromatin organization. Cell Cycle 13: 249-267.
Corpet A., Stucki M. (2014). Chromatin maintenance and dynamics in senescence: a spotlight on SAHF formation and the epigenome of senescent cells. Chromosoma 123: 423-436.
Van Damme E., Laukens K., Dang T. H., Van Ostade X. (2010). A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. International Journal of Biological Sciences 6: 51-67.
Datta N., Chakraborty S., Basu M., Ghosh M. K. (2021). Tumor Suppressors Having Oncogenic Functions: The Double Agents. Cells 10: 46.
Drané P., Ouararhni K., Depaux A., Shuaib M., Hamiche A. (2010). The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes & Development 24: 1253-1265.
Dyer M. A., Qadeer Z. A., Valle-Garcia D., Bernstein E. (2017). ATRX and DAXX: Mechanisms and Mutations. Cold Spring Harbor Perspectives in Medicine 7: a026567.
Ferbeyre G., de Stanchina E., Querido E., Baptiste N., Prives C., Lowe S. W. (2000). PML is induced by oncogenic ras and promotes premature senescence . Genes & Development 14: 2015-2027.
Gaspar-Maia A., Alajem A., Meshorer E., Ramalho-Santos M. (2011). Open chromatin in pluripotency and reprogramming. Nature Reviews Molecular Cell Biology 12: 36-47.
Gialitakis M., Arampatzi P., Makatounakis T., Papamatheakis J. (2010). Gamma Interferon-Dependent Transcriptional Memory via Relocalization of a Gene Locus to PML Nuclear Bodies. Molecular and Cellular Biology 30: 2046-2056.
Goldberg A. D., Banaszynski L. A., Noh K.M., Lewis P. W., Elsaesser S. J., Stadler S., Dewell S., Law M., Guo X., Li X., Wen D., Chapgier A., DeKelver R. C., Miller J. C., Lee Y.L., Boydston E. A., Holmes M. C., Gregory P. D., Greally J. M., Rafii S., Yang C., Scambler P. J., Garrick D., Gibbons R. J., Higgs D. R., Cristea I. M., Urnov F. D., Zheng D., Allis C. D. (2010). Distinct Factors Control Histone Variant H3.3 Localization at Specific Genomic Regions. Cell 140: 678-691.
Gordon D. J., Resio B., Pellman D., (2012). Causes and consequences of aneuploidy in cancer. Nature Reviews Genetics 13: 189-203.
Gupta G. P., Perk J., Acharyya S., de Candia P., Mittal V., Todorova-Manova K., Gerald W. L., Brogi E., Benezra R., Massagué J., (2007). ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proceedings of the National Academy of Sciences 104: 19506-19511.
Gurrieri C., Capodieci P., Bernardi R., Scaglioni P. P., Nafa K., Rush L. J., Verbel D. A., Cordon-Cardo C., Pandolfi P. P. (2004). Loss of the Tumor Suppressor PML in Human Cancers of Multiple Histologic Origins. JNCI Journal of the National Cancer Institute 96: 269-279.
Hadjimichael C., Chanoumidou K., Nikolaou C., Klonizakis A., Theodosi G.I., Makatounakis T., Papamatheakis J., Kretsovali A. (2017). Promyelocytic Leukemia Protein Is an Essential Regulator of Stem Cell Pluripotency and Somatic Cell Reprogramming. Stem Cell Reports 8: 1366-1378.
Hadjimichael C., Chanoumidou K., Papadopoulou N., Arampatzi P., Papamatheakis J., Kretsovali A., (2015). Common stemness regulators of embryonic and cancer stem cells.. World journal of stem cells 7: 1150-1184.
Hayflick L., Moorhead P.S. (1961). The serial cultivation of human diploid cell strains. Experimental Cell Research 25: 585-621.
Hoischen C., Monajembashi S., Weisshart K., Hemmerich P. (2018). Multimodal Light Microscopy Approaches to Reveal Structural and Functional Properties of Promyelocytic Leukemia Nuclear Bodies. Frontiers in Oncology 8: 125.
Hsu K.S., Guan B.J., Cheng X., Guan D., Lam M., Hatzoglou M., Kao H.Y. (2016). Translational control of PML contributes to TNFα-induced apoptosis of MCF7 breast cancer cells and decreased angiogenesis in HUVECs. Cell Death & Differentiation 23: 469-483.
Hsu K.S., Kao H.Y. (2018). PML: Regulation and multifaceted function beyond tumor suppression. Cell & Bioscience 8: 5.
Hsu K.S., Zhao X., Cheng X., Guan D., Mahabeleshwar G. H., Liu Y., Borden E., Jain M. K., Kao H.Y. (2017). Dual regulation of Stat1 and Stat3 by the tumor suppressor protein PML contributes to interferon α-mediated inhibition of angiogenesis. Journal of Biological Chemistry 292: 10048-10060.
Ito K., Bernardi R., Morotti A., Matsuoka S., Saglio G., Ikeda Y., Rosenblatt J., Avigan D. E., Teruya-Feldstein J., Pandolfi P. P. (2008). PML targeting eradicates quiescent leukaemia-initiating cells. Nature 453: 1072-1078.
Ito K., Carracedo A., Weiss D., Arai F., Ala U., Avigan D. E., Schafer Z. T., Evans R. M., Suda T., Lee C.H., Pandolfi P. P. (2012). A PML–PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nature Medicine 18: 1350-1358.
Ivanschitz L., De Thé H., Le Bras M. (2013). PML, SUMOylation, and Senescence. Frontiers in Oncology 3: 171.
Jaenisch R., Young R. (2008). Stem Cells, the Molecular Circuitry of Pluripotency and Nuclear Reprogramming. Cell 132: 567-582.
Jolly M. K., Ware K. E., Gilja S., Somarelli J. A., Levine H. (2017). EMT and MET : necessary or permissive for metastasis? . Molecular Oncology 11: 755-769.
Kakizuka A., Miller W.H., Umesono K., Warrell R.P., Frankel S.R., Murty V.V.V.S., Dmitrovsky E., Evans R.M. (1991). Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor, PML. Cell 66: 663-674.
Kim M. K., Yang S., Lee K.H., Um J.H., Liu M., Kang H., Park S. J., Chung J. H. (2011). Promyelocytic leukemia inhibits adipogenesis, and loss of promyelocytic leukemia results in fat accumulation in mice. American Journal of Physiology-Endocrinology and Metabolism 301: E1130-E1142.
Kumar P. P., Bischof O., Purbey P. K., Notani D., Urlaub H., Dejean A., Galande S. (2007). Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nature Cell Biology 9: 45-56.
Kurihara M., Kato K., Sanbo C., Shigenobu S., Ohkawa Y., Fuchigami T., Miyanari Y. (2020). Genomic Profiling by ALaP-Seq Reveals Transcriptional Regulation by PML Bodies through DNMT3A Exclusion. Molecular Cell 78: 493-505.e8.
Lallemand-Breitenbach V., de Thé H. (2012). Hematopoietic Stem Cells Burn Fat to Prevent Exhaustion. Cell Stem Cell 11: 447-449.
Lallemand-Breitenbach V., de Thé H. (2018). PML nuclear bodies: from architecture to function. Current Opinion in Cell Biology 52: 154-161.
LaMorte V. J., Dyck J. A., Ochs R. L., Evans R. M. (1998). Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proceedings of the National Academy of Sciences 95: 4991-4996.
Lehming N., Le Saux A., Schuller J., Ptashne M. (1998). Chromatin components as part of a putative transcriptional repressing complex. Proceedings of the National Academy of Sciences 95: 7322-7326.
Li W., Ferguson B. J., Khaled W. T., Tevendale M., Stingl J., Poli V., Rich T., Salomoni P., Watson C. J. (2009). PML depletion disrupts normal mammary gland development and skews the composition of the mammary luminal cell progenitor pool. Proceedings of the National Academy of Sciences 106: 4725-4730.
Lin H.K., Bergmann S., Pandolfi P. P. (2004). Cytoplasmic PML function in TGF-β signalling. Nature 431: 205-211.
Liu L. (2017). Linking Telomere Regulation to Stem Cell Pluripotency. Trends in Genetics 33: 16-33.
Luciani J. J, Depetris D., Missirian C., Mignon-Ravix C., Metzler-Guillemain C., Megarbane A., Moncla A., Mattei M.-G., (2005). Subcellular distribution of HP1 proteins is altered in ICF syndrome. European Journal of Human Genetics 13: 41-51.
Luciani J. J., Depetris D., Usson Y., Metzler-Guillemain C., Mignon-Ravix C., Mitchell M. J., Megarbane A., Sarda P., Sirma H., Moncla A., Feunteun J., Mattei M.G. (2006). PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. Journal of Cell Science 119: 2518-2531.
von Mikecz A., Zhang S., Montminy M., Tan E. M., Hemmerich P. (2000). Creb-Binding Protein (Cbp/P300) and RNA Polymerase II Colocalize in Transcriptionally Active Domains in the Nucleus. Journal of Cell Biology 150: 265-274.
Missiroli S., Bonora M., Patergnani S., Poletti F., Perrone M., Gafà R., Magri E., Raimondi A., Lanza G., Tacchetti C., Kroemer G., Pandolfi P. P., Pinton P., Giorgi C. (2016). PML at Mitochondria-Associated Membranes Is Critical for the Repression of Autophagy and Cancer Development. Cell Reports 16: 2415-2427.
Mittal K., Kaur J., Jaczko M., Wei G., Toss M. S., Rakha E. A., Janssen E., Søiland H., Kucuk O., Reid M. D., Gupta M. V., Aneja R., (2021). Centrosome amplification: a quantifiable cancer cell trait with prognostic value in solid malignancies.. Cancer metastasis reviews 40: 319-339.
Morganti C., Missiroli S., Lebiedzinska-Arciszewska M., Ferroni L., Morganti L., Perrone M., Ramaccini D., Occhionorelli S., Zavan B., Wieckowski M. R., Giorgi C. (2019). Regulation of PKCβ levels and autophagy by PML is essential for high-glucose-dependent mesenchymal stem cell adipogenesis. International Journal of Obesity 43: 963-973.
Nisole S., Maroui M. A., Mascle X. H., Aubry M., Chelbi-Alix M. K. (2013). Differential Roles of PML Isoforms. Frontiers in Oncology 3: 125.
Oh W., Ghim J., Lee E.W., Yang M.R., Kim E. T., Ahn J.H., Song J. (2009). PML-IV functions as a negative regulator of telomerase by interacting with TERT. Journal of Cell Science 122: 2613-2622.
Pearson M., Carbone R., Sebastiani C., Cioce M., Fagioli M., Saito S., Higashimoto Y., Appella E., Minucci S., Pandolfi P. P., Pelicci P. G. (2000). PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406: 207-210.
Percherancier Y., Germain-Desprez D., Galisson F., Mascle X. H., Dianoux L., Estephan P., Chelbi-Alix M. K., Aubry M. (2009). Role of SUMO in RNF4-mediated Promyelocytic Leukemia Protein (PML) Degradation. Journal of Biological Chemistry 284: 16595-16608.
Ponente M., Campanini L., Cuttano R., Piunti A., Delledonne G. A., Coltella N., Valsecchi R., Villa A., Cavallaro U., Pattini L., Doglioni C., Bernardi R. (2017). PML promotes metastasis of triple-negative breast cancer through transcriptional regulation of HIF1A target genes. JCI Insight 2: e87380.
Potts P. R., Yu H. (2007). The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nature Structural & Molecular Biology 14: 581-590.
Regad T., Bellodi C., Nicotera P., Salomoni P. (2009). The tumor suppressor Pml regulates cell fate in the developing neocortex. Nature Neuroscience 12: 132-140.
Reineke E. L., Lam M., Liu Q., Liu Y., Stanya K. J., Chang K.S., Means A. R., Kao H.Y. (2008). Degradation of the Tumor Suppressor PML by Pin1 Contributes to the Cancer Phenotype of Breast Cancer MDA-MB-231 Cells. Molecular and Cellular Biology 28: 997-1006.
Reineke E. L., Liu Y., Kao H.Y. (2010). Promyelocytic Leukemia Protein Controls Cell Migration in Response to Hydrogen Peroxide and Insulin-like Growth Factor-1. Journal of Biological Chemistry 285: 9485-9492.
Sachini N., Arampatzi P., Klonizakis A., Nikolaou C., Makatounakis T., Lam E. W.F., Kretsovali A., Papamatheakis J. (2019). Promyelocytic leukemia protein ( PML ) controls breast cancer cell proliferation by modulating Forkhead transcription factors . Molecular Oncology 13: 1369-1387.
Scaglioni P. P., Rabellino A., Yung T. M., Bernardi R., Choi S., Konstantinidou G., Nardella C., Cheng K., Pandolfi P. P. (2012). Translation‐dependent mechanisms lead to PML upregulation and mediate oncogenic K‐RAS‐induced cellular senescence. EMBO Molecular Medicine 4: 594-602.
Schuster B. S., Reed E. H., Parthasarathy R., Jahnke C. N., Caldwell R. M., Bermudez J. G., Ramage H., Good M. C., Hammer D. A. (2018). Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nature Communications 9: 2985.
Shastrula P. K., Sierra I., Deng Z., Keeney F., Hayden J. E., Lieberman P. M., Janicki S. M. (2019). PML is recruited to heterochromatin during S phase and represses DAXX-mediated histone H3.3 chromatin assembly. Journal of Cell Science 132: jcs220970.
Singh A., Denu R. A., Wolfe S. K., Sperger J. M., Schehr J., Witkowsky T., Esbona K., Chappell R. J., Weaver B. A., Burkard M. E., Lang J. M. (2020). Centrosome amplification is a frequent event in circulating tumor cells from subjects with metastatic breast cancer. Molecular Oncology 14: 1898-1909.
Song C., Wang L., Wu X., Wang K., Xie D., Xiao Q., Li S., Jiang K., Liao L., Yates J. R., Lee J.D., Yang Q. (2018). PML Recruits TET2 to Regulate DNA Modification and Cell Proliferation in Response to Chemotherapeutic Agent. Cancer Research 78: 2475-2489.
de Stanchina E., Querido E., Narita M., Davuluri R. V., Pandolfi P. P., Ferbeyre G., Lowe S. W. (2004). PML Is a Direct p53 Target that Modulates p53 Effector Functions. Molecular Cell 13: 523-535.
Stankic M., Pavlovic S., Chin Y., Brogi E., Padua D., Norton L., Massagué J., Benezra R. (2013). TGF-β-Id1 Signaling Opposes Twist1 and Promotes Metastatic Colonization via a Mesenchymal-to-Epithelial Transition. Cell Reports 5: 1228-1242.
Sun J., Fu S., Zhong W., HUANG H. (2013). PML overexpression inhibits proliferation and promotes the osteogenic differentiation of human mesenchymal stem cells. Oncology Reports 30: 2785-2794.
Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007). Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 131: 861-872.
Talluri S., Dick F. A. (2014). The retinoblastoma protein and PML collaborate to organize heterochromatin and silence E2F-responsive genes during senescence. Cell Cycle 13: 641-651.
Tampakaki M., Oraiopoulou M.E., Tzamali E., Tzedakis G., Makatounakis T., Zacharakis G., Papamatheakis J., Sakkalis V. (2021). PML Differentially Regulates Growth and Invasion in Brain Cancer. International Journal of Molecular Sciences 22: 6289.
Tang H., Jin Y., Jin S., Tan Z., Peng Z., Kuang Y. (2016). Arsenite inhibits the function of CD133+ CD13+ liver cancer stem cells by reducing PML and Oct4 protein expression. Tumor Biology 37: 14103-14115.
de Thé H., Lavau C., Marchio A., Chomienne C., Degos L., Dejean A. (1991). The PML-RARα fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 66: 675-684.
Trotman L. C., Alimonti A., Scaglioni P. P., Koutcher J. A., Cordon-Cardo C., Pandolfi P. P. (2006). Identification of a tumour suppressor network opposing nuclear Akt function. Nature 441: 523-527.
Tsai J. H., Donaher J. L., Murphy D. A., Chau S., Yang J. (2012). Spatiotemporal Regulation of Epithelial-Mesenchymal Transition Is Essential for Squamous Cell Carcinoma Metastasis. Cancer Cell 22: 725-736.
Udroiu I., Sgura A. (2020). Alternative Lengthening of Telomeres and Chromatin Status. Genes 11: 45.
Uversky V. N. (2017). Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Current Opinion in Structural Biology 44: 18-30.
Vernier M., Bourdeau V., Gaumont-Leclerc M.F., Moiseeva O., Bégin V., Saad F., Mes-Masson A.M., Ferbeyre G. (2011). Regulation of E2Fs and senescence by PML nuclear bodies. Genes & Development 25: 41-50.
Wang J., Shiels C., Sasieni P., Wu P. J., Islam S. A., Freemont P. S., Sheer D. (2004). Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. Journal of Cell Biology 164: 515-526.
Wang Z.G., Ruggero D., Ronchetti S., Zhong S., Gaboli M., Rivi R., Pandolfi P. P. (1998). Pml is essential for multiple apoptotic pathways. Nature Genetics 20: 266-272.
Wong L. H., McGhie J. D., Sim M., Anderson M. A., Ahn S., Hannan R. D., George A. J., Morgan K. A., Mann J. R., Choo K.H. A. (2010). ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Research 20: 351-360.
Yamada N., Tsujimura N., Kumazaki M., Shinohara H., Taniguchi K., Nakagawa Y., Naoe T., Akao Y. (2014). Colorectal cancer cell-derived microvesicles containing microRNA-1246 promote angiogenesis by activating Smad 1/5/8 signaling elicited by PML down-regulation in endothelial cells. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1839: 1256-1272.
Yang L., Shi P., Zhao G., Xu J., Peng W., Zhang J., Zhang G., Wang X., Dong Z., Chen F., Cui H. (2020). Targeting cancer stem cell pathways for cancer therapy. Signal Transduction and Targeted Therapy 5: 8.
Zhang R., Chen W., Adams P. D. (2007). Molecular Dissection of Formation of Senescence-Associated Heterochromatin Foci. Molecular and Cellular Biology 27: 2343-2358.
Zhang R., Poustovoitov M. V., Ye X., Santos H. A., Chen W., Daganzo S. M., Erzberger J. P., Serebriiskii I. G., Canutescu A. A., Dunbrack R. L., Pehrson J. R., Berger J. M., Kaufman P. D., Adams P. D. (2005). Formation of MacroH2A-Containing Senescence-Associated Heterochromatin Foci and Senescence Driven by ASF1a and HIRA. Developmental Cell 8: 19-30.
Zhu J., Zhou J., Peres L., Riaucoux F., Honoré N., Kogan S., de Thé H. (2005). A sumoylation site in PML/RARA is essential for leukemic transformation. Cancer Cell 7: 143-153.